Local Phosphate Sensing
We are using complementary approaches in Arabidopsis thaliana to dissect plant phosphate (Pi) deficiency responses at multiple levels, with an emphasis on local root Pi sensing. These approaches range from mutant isolation and characterization to the analysis of gene and protein function as well as their underlying regulatory networks. We employ forward, reverse, and molecular genetics, biochemistry and structural biology, multi-omics and bioinformatics, as well as cell biology. Our studies of PDR (PHOSPHATE DEFICIENCY RESPONSE), LPR (LOW PHOSPHATE ROOT), and PDR SUPPRESSOR genes provided first insights and models of local root Pi sensing in the context of antagonistic interactions between Pi and associated metallic cations.
Phosphate - Metal Interactions
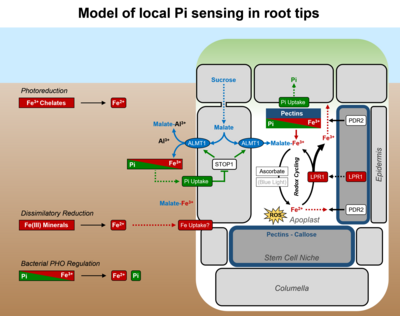
Inhibition of primary root growth by Pi deprivation proceeds via rapid cessation of cell elongation (<2 h) in the transition zone, followed by progressive attenuation of cell division (<2 d) in the root apical meristem. Remarkably, the response of root tips to low Pi availability requires presence of external Fe, which suggests monitoring of Pi via Pi-metal interactions (for review, see Abel 2017).
Two interacting genes are key in local root tip Pi sensing. PDR2 codes for the single ER-resident P5-type ATPase, ATP5A, which functions in ER stress response (Ticconi et al. 2009) and ER stress-dependent autophagy (Naumannn et al. 2019). PDR2 is thought to either restrict LPR1 secretion, a cell wall-targeted ferroxidase, or to limit LPR1 reactant (Fe2+ / Fe3+) availability in the apoplast. In low Pi, the LPR1 expression domain determines cell type-specific Fe accumulation in cell walls of the root apex (Müller et al. 2015). Interestingly, LPR1 typifies an ancient cohort of Fe-oxidizing multicopper oxidases (MCOs) of bacterial origin, regulated by Fe availability (Naumann et al. 2022). A pdr2 suppressor screen identified additional lpr1 mutations (supporting the functional interaction of both gene loci) and two new genes: ALMT1 (ALUMINUM ACTIVATED MALATE TRANSPORTER 1) and STOP1 (SENSITIVE TO PROTON RHIZOTOXICITY 1). Both genes interact to activate malate exudation into the rhizosphere and cell wall space upon Pi limitation. ALMT1 is a direct target gene of the STOP1 transcription factor and encodes a malate efflux channel of key importance for Al detoxification. Chemical (malate) complementation of the almt1 root phenotype on low Pi points to intricate Pi-Al interactions and to a critical role of metal (Fe, Al) chelation in root Pi sensing (Balzergue et al. 2017).
Antagonistic Pi-Fe interactions in the apoplast may determine LPR1-dependent Fe redox cycling, ROS formation, callose deposition, and symplastic connectivity via plasmodesmata to adjust root cell elongation and root meristem maintenance in response to external Pi status (Müller et al. 2015, Naumann et al. 2022).
The interaction of PDR2 and LPR1 together with ALMT1 and STOP1 provides a bridgehead from which to explore local Pi sensing. We are further pursuing the following topics:
- Biochemical function and regulation of LPR1-type ferroxidases
- Regulation of Fe redox cycling by apoplast chemistry
- Pi/Fe-dependent redox signaling in local Pi response
- Interaction of Pi deficiency and associated metal toxicities (Al, Fe)
Evolution of Local Pi Scouting in Land Plants
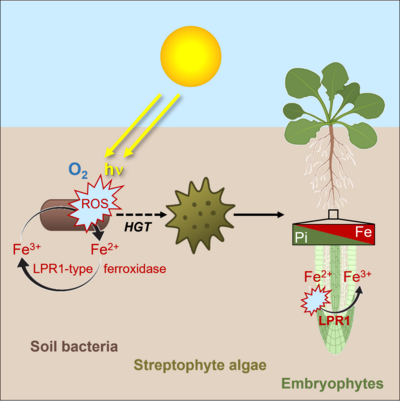
The progenitor of streptophytes (charophyte algae and embryophytes) acquired the LPR1-type MCO ferroxidase via an horizontal gene transfer (HGT) from soil bacteria (Naumann et al. 2022, Abel and Naumann 2024). Thus, LPR1-type MCOs provide an ideal experimental system to investigate the effects of HGT on early land plant evolution and their adaptations to geochemical conditions in terrestrial habitats. Here, we investigate the biochemical development of LPR1-like ferroxidases from soil bacteria, mosses, ferns and vascular plants with respect to their enzymatic activity, substrate specificity and enzyme kinetics. Within the DFG Priority Program 2237 (MAdLand - Molecular Adaptation to Land: plant evolution to change) we study the evolution of LPR1-like enzyme specificity and selectivity in bacteria and the model system Marchantia polymorpha to explore the impact of HGT on the evolution of early local Pi sensing during the water-to-land transition, when plants faced dramatically different geochemical conditions.
In the SFB 1664 (Plant Proteoform Diversity: Bridging the gap from genetic to phenotypic variation) in collaboration with the MLU, we aim to identify direct LPR1 interactors (e.g. Fe transporters) in Arabidopsis accessions (adapted to various soil conditions and climate factors) and determine the enzymatic properties associated with ecotype-specific LPR1 proteoforms resulting from single nucleotide polymorphisms.
Plant P5A-type ATPases- Guardians of ER Quality Control
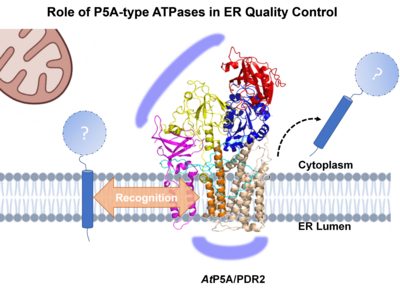
While LPR1 ferroxidase activity monitors Pi availability in root tips, PDR2 counteracts LPR1 by maintaining root tip Fe homeostasis. Recently published biochemical and genetic studies on PDR2/AtP5A orthologs in yeast (Spf1p), nematodes (CATP-8) and human cell lines (ATP13A1) point to a central role of P5A-ATPases as transmembrane helix dislocase, which is thought to enforce quality control by disposing mislocalized or misinserted proteins from the ER membrane to allow for another attempt of correct protein localization and thus to prevent premature protein degradation. However, the spectrum of P5A clients, regulatory interactors in the local ER environment, and the molecular mode of target recognition are unknown. In contrast to the severe (often embryo lethal) and pleiotropic phenotypes in Mammalia, loss of plant P5A-ATPase causes Pi-conditional and organ-specific defects, thus providing an ideal experimental system for the study of P5A function. Accordingly, Pi limitation induces ER stress-mediated autophagy (Naumann et al. 2019, Stephanie et al. 2020, Picchianti et al. 2023) as a subsequent, terminal process of ER quality control via the PDR2-LPR1 interaction, suggesting a direct link between early ER quality control, ER protein homeostasis and adaptation of root development to Pi availability. Our research focuses on the investigation of:
- cell biological function of PDR2/AtP5A in Pi-dependent Fe homeostasis
- mechanistic action of PDR2/AtP5A in ER quality control
- role of PDR2/AtP5A at organellar membrane contact sites
- regulation of biochemical AtP5A/PDR2 function
- function of ER stress-mediated autophagy in local Pi sensing
This page was last modified on 27 Jan 2025 25 Apr 2025 25 Apr 2025 25 Apr 2025 25 Apr 2025 .