Biosynthesis und Function of pollen-specific phenolamides und flavonolglycosides
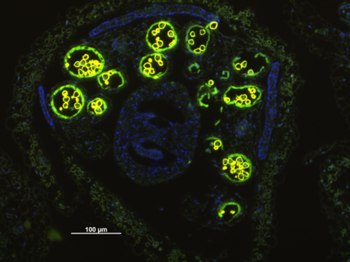
Fig. 1: Cross-section through an Arabidopsis anther. The tapetal expression of the glucosyltransferase UGT79B6 is shown by fusion of the UGT79B6 promotor to GFP and subsequent immunolabelling with an anti-GFP polyclonal antibody visualized by a secondary antibody conjugated to Alexa Fluor 488 resulting in a greenish fluorescence of the tapetum surrounding the autofluorescing pollen grains.
Like pollen of most angiosperms, Arabidopsis accumulates two types of phenylpropanoids: flavonoldiglycosides (sophorosides) and trishydroxycinnamic acid-spermidin-conjugates (phenolamides) on the pollen surface. Besides the structural identification of these pollen-specific conjugates, the localization of the corresponding tapetum specific enzymes (Figure 1), and the annotation of tapetum-localized biosynthetic pathways we currently focus on the transport of these compounds to the pollen surface. Comparison of the modified phenylpropanoid pattern in the tryphine of putative Arabidopsis transporter mutants versus pollen from wildtype enables the selection of transporter candidates and will allow conclusions on transport of phenolamides and flavonols, versus deposition based on apoptosis of the tapetum. Although both types of compounds are universally present on the pollen of angiosperms, investigations on one or several possible in vivo function have not been conclusive yet. From protection against UV-light, enhanced pathogen resistances towards pollen specific microbes, to increased structural fortification of the pollen wall or enhanced vitality of the germinating pollen, several scenarios appear still plausible.
Biosynthesis of piperine and piperamides in black pepper (Piper nigrum)
Within this project we currently address the biosynthetic pathway of piperine and piperamide conjugate formation in fruits of a non-model species, commercially used black pepper (Piper nigrum). Piperamides are a characteristic subtype of hydroxycinnamic acid amides, which can also be classified as piperidine alkaloids, responsible for the pungent taste of this widely used spice. A combination of classical organic synthesis and enzymology with state of the art molecular tools of RNA-Seq analysis should enable us to clarify the various unknown and potentially unique steps of this metabolic grid, resulting in the formation of the phenylpropanoid piperic acid and the amino acid derived piperidine heterocycle. Piperine synthase catalyzing piperine formation from piperoyl CoA and piperidine (Figure 2) as well as the methylene dioxybridge within the aromatic part of the molecule are currently investigated. Based on classical enzyme purification in the case of piperine synthase, database mining, and comparing RNA-seq profiles of fruits high in piperine with leaves, flowers, and roots, all low in piperine, first candidate transcripts have been obtained and are currently investigated for substrate profile and specificity. Besides the aromatic part of the piperine molecule the molecular approach also resulted in the identification of candidate genes essential for the biosynthesis of the piperidine heterocycle present in many important alkaloids, such as coniin and lobelin.
This page was last modified on 27 Jan 2025 04 Feb 2025 27 Jan 2025 .


