Exciting new enzymes and reactions
As the directed evolution requires several cycles and the expression of thousands of different mutants, most protein engineering campaigns rely on E. coli as a simple expression host. However, this focus on a prokaryotic host restricts the enzyme diversity. Several animal, plant, and fungus-derived enzymes are post-translationally modified and not or barely producible in E. coli.
The yeast Pichia pastoris can perform post-translational modifications and has proven to secrete proteins on an industrial-relevant scale. We developed a modular secretion and detection system, enabling the screening of large enzyme libraries in Pichia pastoris. The screening was performed with an exciting C–H activating enzyme class: the unspecific peroxygenases (UPOs). An enzyme class, which can perform hydroxylation reactions of non-activated sp3-hybridized carbons. However, only one variant could be heterologously produced in yeast when we started the project.
Modular yeast secretion technique
We developed a Golden Gate-based system, which enabled the rapid screening of different parts of an expression/secretion cassette and started with Saccharomyces cerevisiae as the more established yeast organism. This cassette consisted of a signal peptide responsible for the secretion, different UPO genes, and a C-terminal-tag responsible for protein purification and detection via split-GFP. With the Golden Gate principle, we were able to identify the ideal C-terminal tag and the best signal peptide/UPO gene combination and were able to secrete seven out of the tested eight UPOs (Commun. Biol. 4, 2021)! All developed plasmids are available at AddGene.
Directed Evolution
of Unspecific Peroxygenases, UPOs
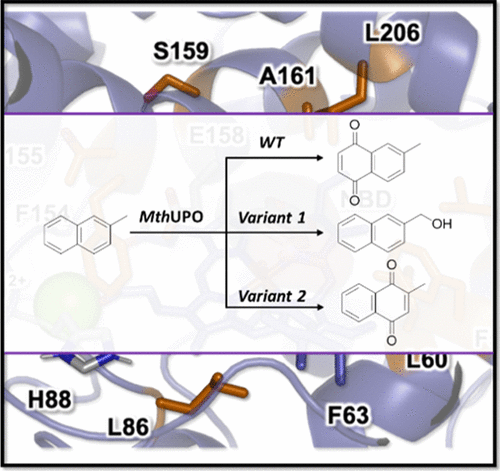
With the new system to access and screen new UPOs, we utilized the MthUPO from a thermophilic fungus to perform directed evolution. The aim was to create chemodivergent variants, where one can hydroxylate at the benzylic position and the other oxyfunctionalizes the aromatic ring (ACS Catal. 11, 2021). Initial single site-saturation libraries were created using Golden Mutagenesis. All hits were recombined in an extensive library. The screening of more than 5000 transformants resulted in the identification of the selective variants. The aromatic oxyfunctionalization produces the important compound vitamin K3 (collaboration with Marc Garcia-Borras).
Expansion of the system to Pichia pastoris
To adapt the system to Pichia pastoris, an episomal and integrative vector P. pastoris vector was designed. The episomal system has higher transformation efficiency, less interclonal variability, and easier sequencing, whereas the integrative system is more stable. In P. pastoris, by far, most work has been done using genomic integration concepts. Both systems were tested, resulting in the successful secretion of a range of UPOs using two different promoters.
Preparative scale UPO conversion
The new P. pastoris system resulted in improved production titers of several UPOs. With these titers, sufficient amounts of the enzyme could be isolated to perform preparative scale conversion of important phenethylamine-derivatives and isolate the enantioselectively hydroxylated product (Commun. Biol. 4, 2021).
Promoter screening in Pichia pastoris
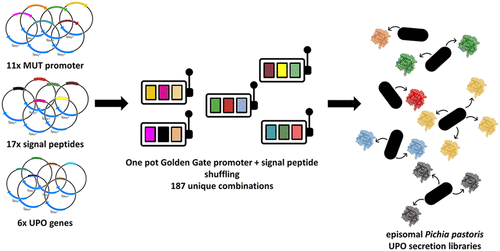
The previously developed episomal expression system in P. pastoris has the advantage that large libraries can be screened. We noticed that the combination of the promoter and the signal peptide has a tremendous and not predictable influence. We expanded the tripartite system with a fourth module: promoters. 11 different promoters from P. pastoris and Hansenula polymorpha were screened in combination with the 17 different signal peptides. The screening resulted in several cases of substantially improved secretions for UPOs, lipases, and laccases. Also, the secretion of three further UPOs was described for the first time in yeast (ACS Synth. Biol. 10, 2021). All developed plasmids are available at AddGene.
This page was last modified on 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 .