Please visit our external website for up-to-date information: dissmeyerlab.org
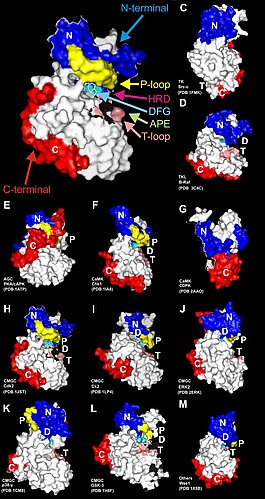
Figure 1: Crucial factors for the function of proteins are their concentrations and three-dimensional structures. The figure shows different three-dimensional structures of highly related biocatalytic active proteins (enzymes). In this case kinases are shown, which can e.g. bind phosphate to other proteins in order to change their properties by regulating their activities. For further information read the current review article (Dissmeyer & Schnittger (2011), The age of protein kinases, Methods Mol Biol. 2011;779: 7-52 (free download)
Proteins belong to the fundamental equipment of each cell of all living organisms. In general, proteins participate in virtually every cellular process and play vital roles as catalysts and controllers in numerous biochemical reactions, as well as structural scaffolds of the cell’s architecture. They are only able to function properly if their abundance and shape – with respect to stability and three-dimensional folding – are correct.
Thus, understanding the factors behind protein stability and folding has a wide-spread importance in molecular biology. Therefore, we need to know what the molecular mechanisms of recognition and degradation of erroneous or destabilized proteins are.
Proteins consist of linked building blocks (amino acids) in a particular order resulting in a specific three-dimensional shape, the so-called fold. The overall entity of proteins, the proteome, must be precisely controlled to function properly. This is come along by transcriptional, translational, and posttranslational control on the level of protein abundance (proteostasis) but also by protein quality control (PQC) mechanisms sensing erroneous and misfolded proteins. Therefore, the cells host different quality control mechanisms with checkpoint function. When proteins are wrongly assembled, have an incorrect shape or are "used up" after their action, they have to be removed from the cell by proteolysis. Also "non-stop" proteins that remain stuck in the ribosome, the protein manufacturing plant of the cell, must be sensed and destroyed.
Specialized protein quality control systems as parts of the ubiquitin proteasome system (UPS), are responsible for essential targeted proteolysis and removal of either damaged polypeptides or proteins that harbor specific destruction tags. Despite their clear involvement in crucial cellular functions, some but not many enzymatic components of these pathways have been identified.
We want to further elucidate the composition of the pool of in vivo substrates of the proteolysis networks to be able to describe their biological roles and the processes they are involved. That requires more insight into the molecular function of protein quality "checkpoints" and how cells sense and survive polypeptides that need to be destructed. Furthermore, it is vital to know how protein abundance can be controlled in physiological contexts. The integrated research program suggested here will shed new light on understanding and possibly improving environmental stress tolerance in plants. This, in turn, has direct implications on functional plant proteins representing the premier storage unit for energy needed in form of food and biofuels.
We have developed an in vivo transgenic protein stability reporter system that allows screening for mutants defective in protein quality control pathways. As model system, we are using Arabidopsis, currently the best understood plant organism, by combining state-of-the-art genetics and cell biology with high-tech biochemistry techniques as experimental approaches. Key aims of our research are to understand the intricate functional network of protein quality control in plants.
This page was last modified on 27 Jan 2025 27 Jan 2025 27 Jan 2025 .

