Methodology
Identification of Enzymes
The application of biocatalytic reactions in chemical syntheses is often limited due to a lack of suitable enzymes. Therefore, the initial step of our research is to identify enzyme candidates for a desired reaction by mining genome or transcriptome data. These are increasingly available for plants and fungi which produce natural products (NPs) of a class of NPs of interest. In addition, we also screen for enzymes directly, e.g. from the producer organisms of our natural product target of interest or related species. Enzymes with a naturally wide-range substrate spectrum are preferred ("promiscous enzymes, see Fig. 1). All studies for the discovery of new or suitable enzymes are closely related to the development of enzymatic assays.
Figure1:
Implementation of non-natural substrates using a coenzyme A ligase with a wide substrate range. The plant enzyme converts carboxylic acids into “activated” coenzyme A thioesters (a), which in turn are the basis for a large number of biosyntheses (polyketides, flavonoids, coumarines etc.). The enzyme identified here can be applied in many ways, since in addition to the natural substrates (marked in red) it also converts many other structurally different substrates (b) (Dippe et al., Adv. Synth. Cat. 2019, 361 (23), 5346-5350. DOI: 10.1002/adsc.201900892).
Optimization of Biocatalysts
Some properties of enzymes obtained from natural sources limit their wider applicability in lab or industry. Examples are low or no activity towards non-natural substrates, lack of regiospecificity, substrate or product inhibition. In order to improve such disadvantageous properties, we use structure-based optimization methods such as rational re-design or homology modelling based methods for predicting advantageous mutations (see Fig. 2, work together with RG Computational Chemistry). E.g., optimizations of cofactor-forming enzymes improve access and range of these cost-limiting substances important for many enzyme classes to work properly (methyl transferases, coenzyme A ligases, hydroxylases).
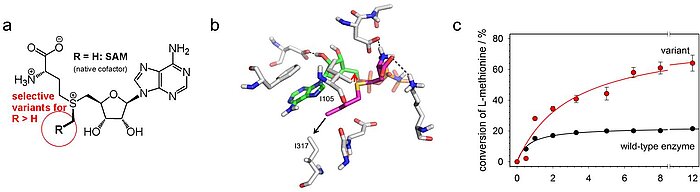
Figure 2: Rational re-design of an S-adenosylmethionine synthetase (according to Dippe et al., Chem. Commun. 2016). This enzyme provides the cofactor SAM (a, R = H), which is used in almost all biological methylation reactions, but is limited by product inhibition. An exchange of amino acid residues in the enzymes active center (b) leads to enzyme variants which are suitable for the high-yield synthesis of SAM without product inhibition (c). The variants can also produce non-natural SAM derivatives R > H (a).
Enzyme Applications
Our research primarily focuses on multi-enzyme cascades, i.e. sequential enzyme-catalyzed reactions that produces a certain natural bioactive compound or intermediate from simple precursor molecules. Purified enzymes are used, for example, in an immobilized form for catalysis. In this in vitro method toxic compounds can be used and produced, e.g. antibiotics, and it can be combined with chemical steps and run in standard chemical reactors (drop-in technology). Usually also concentrations of compounds are higher and purification of the products is simpler than in organism based systems.
An alternative, are transformations of whole (artificial) pathways into host microorganisms (whole-cell biocatalysts), i.e. the enzyme cascade is produced in vivo ("synthetic biology" SynBio). The starting substance is added to the cell suspension, taken up by the organism, and ideally transformed into the product. Two of several advantages of an in vivo production system are that cofactors and standard building blocks are provided by the host organism, and that sensitive intermediates and enzymes are easier to deal with.
The products can be obtained by extraction. For the production of the bitter-masker (taste modifier) homeriodictyol shown in Fig. 3 we could provide both alternatives to an industrial partner.
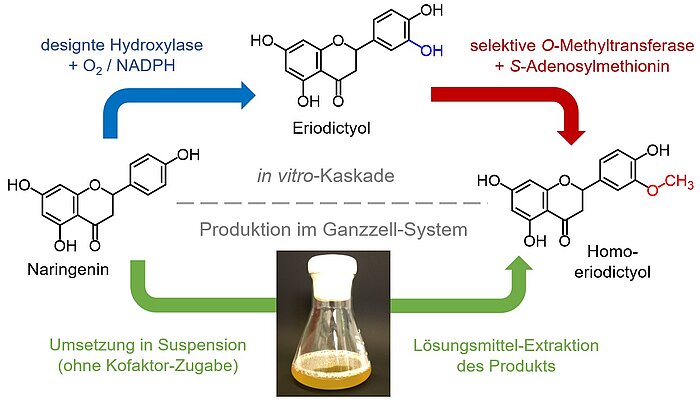
Figure 3: Biocatalytic conversion of the citrus peel waste derived naringenin into the high-price bitter-masker homoeriodictyol (HED). The cascade reaction contains an optimized hydroxylase and a regioselective O-methylation enzyme. The process can be carried out as a cell-free synthesis (top), or intracellularly in recombinant bacteria (bottom) (patent WO2016 / 050656A1, patent application 15188136.4). In the latter process, the required cofactors are provided by the host metabolism, while additional regeneration enzymes are present in the cell-free system.
This page was last modified on 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 .


