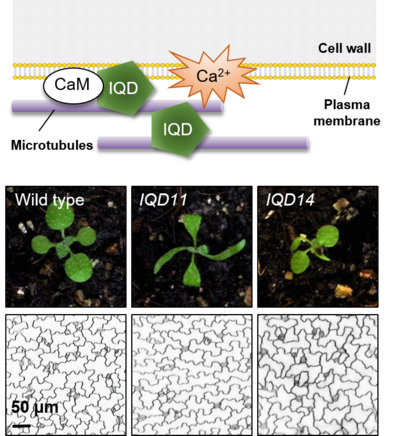
Plant-specific IQD gene families have been comprehensively annotated in the genomes of several angiosperms and encode about 30 predicted IQD proteins in Arabidopsis, rice, and tomato (reviewed in Kölling et al., 2019). The presence of IQD genes in Physcomitrella patens but absence in algae suggests that IQD proteins are an ancient family of CaM/CML target proteins that originated during the early evolution of land plants, possibly before the divergence of bryophyte and vascular plant lineages. The common feature of IQD proteins is the presence of a central region of 67 conserved amino acid residues, referred to as the IQ67 domain, which is necessary and sufficient for CaM binding. Outside of the CaM binding domain, IQD proteins are predicted to contain large regions of intrinsic disorder, which is a hallmark of scaffold proteins embarked in the assembly of macromolecular complexes. A comprehensive characterization of the entire Arabidopsis IQD family revealed preferential localization to microtubules, the plasma membrane and inside the nucleus (Bürstenbinder et al., 2017). Interaction studies in yeast two-hybrid assays identified Kinesin Light Chain-Related (KLCR) proteins as binding partners of IQD1 and related proteins (Bürstenbinder et al., 2013). Coexpression of GFP-IQD1 with RFP-CaM2 and RFP-KLCR in transient expression assays in Nicotiana benthamiana mediates IQD-dependent microtubule recruitment of CaMs and KLCRs. Collectively, we propose that IQDs serve as scaffolds for the assembly of CaM signaling modules to link calcium signaling to the regulation of the microtubule cytoskeleton during plant growth regulation.
Project 1: Function of IQD proteins in plant growth regulation
To investigate the biological function of IQDs and members of the interacting KLCR family, we are generating a platform of IQD and KLCR family resources using reverse genetics. Phenotypes observed in IQD gain-of-function alleles point to the involvement of IQD proteins in regulation of microtubule organization and various processes related to plant development (Bürstenbinder et al., 2017). Analysis of PromoterIQD:GFP-GUS lines in terms of spatio-temporal expression domains revealed preferential promoter activities of most IQD family members in actively dividing and/or expanding tissues (Bürstenbinder et al., 2017; Mitra et al., 2019). Informed by the expression analysis, we screened Arabidopsis iqd loss-of-function lines for phenotypes in tissues with endogenous IQD expression. First available data from analysis of knockout lines link IQD function to regulation of microtubule organization and cellulose deposition during morphogenesis of leaf epidermis pavement cells (Mitra et al., 2019). Current projects extend the phenotypic characterization of iqd and klcr knock out lines by molecular and cell biological as well as physiological analyses. Central aspects of interest are:
- Function of IQDs at the microtubule cytoskeleton
- IQD-mediated regulation of cell wall biosynthesis and composition
- Identification and characterization of IQD-interacting proteins
- Calcium-CaM mediated regulation of IQD function
- Role of IQDs as hubs in cellular signaling (calcium, phytohormones, kinase signaling)
Project 2: Structure-function studies and CaM-mediated regulation of IQD proteins
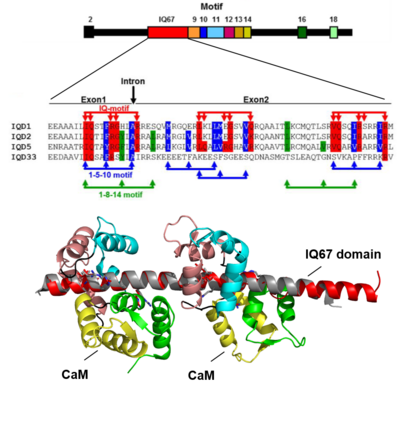
IQDs comprise the largest known class of CaM target proteins in plants. The eponymous IQ67 domain is characterized by the invariant spacing of up to three IQ motifs separated by 11 and 15 intervening residues, which mediate calcium-independent CaM binding. The predicted IQ motifs partially overlap with one to four copies of the 1-5-10 as well as 1-8-14 motifs that are implicated in calcium-dependent CaM binding. In vitro assays with recombinant IQD proteins revealed binding of several IQDs to calcium-free apoCaM and to calcium-bound holoCaM, indicating functionality of the distinct classes of CaM binding motifs (Bürstenbinder et al., 2013; Mitra et al., 2019). Although IQDs are structurally diverse with respect to their computed molecular mass (12-90 kDa), they are, with only a few exceptions, quite uniform at the physicochemical level such as basic isoelectric points (pI~10.3) and a high content of regions with predicted intrinsic disorder. The regions of predicted intrinsic disorder are interspersed by short and family-specific motifs (Abel et al., 2013; Bürstenbinder et al., 2017). In Arabidopsis, IQD proteins group into four distinct phylogenetic classes that differ mostly in the presence and distribution of IQD-specific motifs. Distinct motifs may reflect specific functions of the individual phylogenetic groups, e.g., by providing specificity in protein-protein interactions. First analyses of N- and C-terminal truncated variants of IQD proteins in terms of subcellular localization points to important roles of polybasic stretches in microtubule-binding and plasma membrane association, possibly via electrostatic interactions between acidic tails of tubulin-microtubule subunits or negatively charged headgroups of membrane lipids. Recent work aims at elucidating mechanisms underlying interactions between IQDs, CaMs, KLCRs and additional interacting proteins (Bürstenbinder et al., 2013) to gain insights into the mode and regulation of IQD protein function. Collectively, insights into the biochemical functions and biological roles of IQD proteins will help to unravel complex calcium signaling networks in plants. Topics covered in this project are:
- Structure analysis of select IQD proteins
- Mapping of CaM-binding sites and stoichiometry of IQD-CaM complexes
- Role of CaM in interaction of IQDs with other proteins
- Characterization of IQD-specific motifs in protein-protein interaction and as determinants for subcellular localization
- Regulation of IQD function by posttranslational modifications
Project 3: Development of bioinformatics tools for automatic image analysis
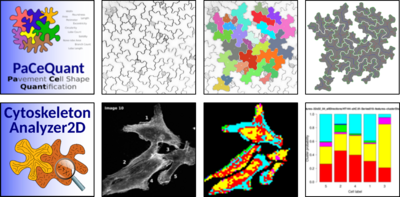
Our cell biological assays frequently generate large amounts of image material that are the basis for comparative analyses. To facilitate robust, unbiased, and high-throughput image analysis and statistical evaluation we collaborate with computer scientists and bioinformaticians at the MLU Halle-Wittenberg and the German Center for Integrative Biodiversity Research (iDiv) to develop bioinformatics tools. We aim to generate user-friendly open source tools for batch processing of large image data sets, which are released in the frame of the ImageJ plugin Microscopy Image Analysis Toolbox (MiTiBo, http://mitobo.informatik.uni-halle.de). For statistical evaluation and graphical visualization we provide supplemental R scripts. So far, we have released PaCeQuant, a tool for automatic quantification of cell shapes (1, 2), and CytoskeletonAnalyzer2D, which enables quantification of global microtubule patterns in complex shaped cells (3, 4, 5). Recent projects involve:
- Optimization and extension of available tools
- Development of novel tools and novel functionalities (as needed)
References to project 3
1 Möller B*, Poeschl Y, Plötner R, Bürstenbinder K.* (2017) PaCeQuant: A tool for high-throughput quantification of pavement cell shape characteristics. Plant Physiol 175(3): 998-1017.
3 Bürstenbinder K*, Möller B, Plötner R, Stamm G, Hause G, Mitra, D, Abel S. (2017) The IQD family of calmodulin binding proteins links calcium signaling to microtubules, membrane subdomains, and the nucleus. Plant Physiol 173:1692-1708.
5 Möller B, Poeschl Y, Klemm S, Bürstenbinder K.* Morphological analysis of leaf epidermis pavement cells with PaCeQuant. Springer. Plant Cell Morphogenesis – Methods and Protocols 2nd edition.
This page was last modified on 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 .