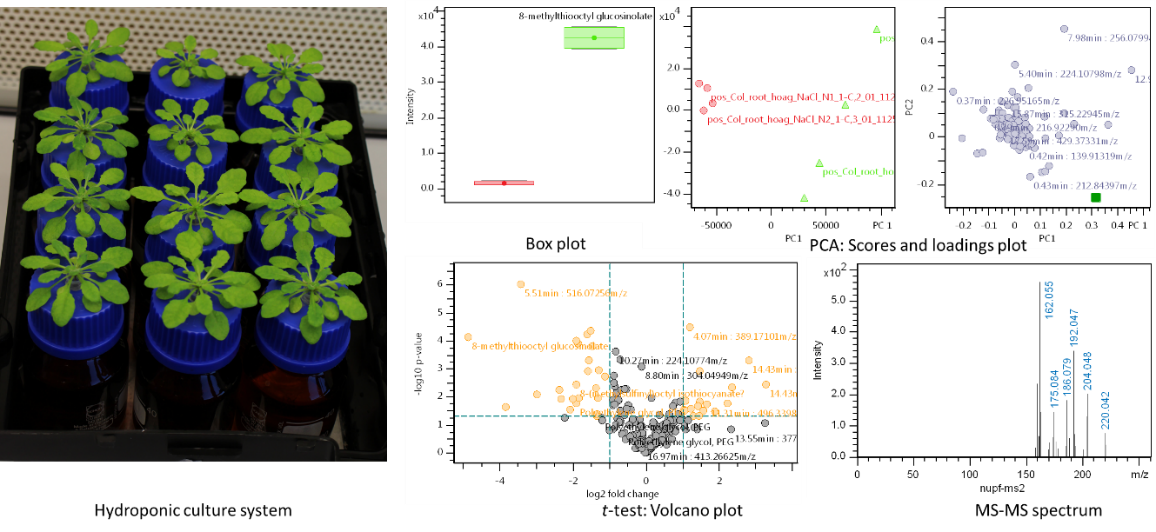
Metabolite profiling of Arabidopsis thaliana during various types of stress
Plants have the remarkable ability to produce more than 100,000 low-molecular-mass compounds, which are called secondary metabolites. Most of these are considered as non-essential for basic metabolism, while many appear to have evolved as components of the plant's response to stress. For many scientific questions aiming to understand the underlying mechanisms it is most important to get an overview of the metabolite profile of the investigated plant, in order to compare the metabolome before and after application of stress. The model plant Arabidopsis thaliana with its sequenced and largely annotated genome and its vast collection of mutant alleles not only provides a convenient system for the study of stress events, it also allows the direct analysis of gene functions and regulatory mechanisms. Although not being a crop or pharmacologically used plant, most of the main pathways are fully developed and many can be activated by stress.
We use several experimental setups (as hydroponics or in climate chambers) to elicit the production of secondary compounds in Arabidopsis. A standardised workflow including sampling, sample preparation, separation by LC, and measurement by ESI-ToF-MSn along with several procedures for quality control enables high reproducibility of results and the application of efficient methods for data analysis.
Our untargeted profiling spans feature extraction and alignment, normalisation, adduct grouping and relative quantification. These data are then ready for Principle Component Analysis, comparison of mean values and feature correlation. Thus, relevant features can be subjected to further analysis like substance identification and absolute quantification.
We continuously extend our metabolite atlas of Arabidopsis for automated annotation, which currently contains several hundreds of secondary compounds from the phenylpropanoids, Brassicaceae-specific glucosinolates, phytoalexins and indoles along with several fatty acid and amino acid derivatives. Our compound library is currently extended by an in-house spectral library.
13C-Isotope-labeling of Arabidopsis thaliana: Metabolic links between photosynthesis, metabolite transport, turnover and exudation
Plants interact with their belowground environment by the secretion of organic compounds called exudates. The metabolic origin of these compounds, meaning the dynamics between carbon fixation, transport, metabolism and exudation is still hardly understood. The application of 13CO2 instead of the normal atmospheric 12CO2 labels metabolites downstream of photosynthesis. The labeled metabolites appear heavier in the mass spectrum (Fig. 2, lower panel) and peak intensity ratios reveal conclusions about the metabolite labeling status. Time-course harvesting and compartmentalization allow the description of tissue-specific metabolite abundances, metabolic turnover, labeling rates as well as transportation and exudation processes.
Relationship of biodiversity and exuded as well as inner root metabolites in grassland communities
While much research has been done on the field of plant metabolomics under laboratory conditions, there is still a blind spot on the behaviour of plants in the rhizosphere under natural conditions and what kind of contribution exudates have to shape the biodiversity of this habitat. This DFG-funded cooperative project between ecology and metabolomics aims to fill this gap by the application of untargeted metabolite profiling on exudates and inner root metabolites, as well as ecological methods of two common grassland life forms: grasses and forbs. Ten different species of these life forms were grown in the German Biodiversity Exploratories (DFG priority program) under natural conditions (field) as well as in a phytochamber under controlled conditions (laboratory). By taking advantage of a grinding robot and both mass spectrometric approaches, liquid and gas chromatography-coupled mass spectrometry, samples of exudates and roots of both experimental set ups are analysed for their primary and secondary metabolite profiles. Metabolites are identified with the help of databases and advanced MS approaches, e.g. MSn, GC-APCI-TOF-MS and H/D exchange. Different bioinformatical and statistical analyses of all four data sets, exuded and root metabolite profiles in field and laboratory, serve to investigate if
(i) plant exudation behaviour is driven by the plant family or
(ii) the species itself. In cooperation with ecologists and microbiologists as well as combination of field and laboratory results this research will also help to understand
(iii) the functional role of exuded root metabolites in the biodiversity of the rhizosphere and
(iv) whether results from the laboratory could be retrieved in a natural environment. Furthermore, the comparison of root and exudates results will help to reveal if
(v) root metabolism can reflect the exuded profile.
This page was last modified on 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 27 Jan 2025 .