IPB researchers Tom Schreiber and colleagues from the Tissier Lab recently developed a two-component AND-gate system for synthetic biology applications in plants. Gates – known rather in context of information technology than biology – are conditional switches. The condition for such a switch to be activated can be ‘AND’. For a two component system, it means, that both components (component 1 ‘AND’ component 2) are required for a positive output to occur. Such a two-component AND-gate can be very helpful in biological systems, e.g., to activate the expression of a specific gene upon a bipartite cue.
Schreiber et al. base their system on transcription activator-like effectors, TALEs for short, which are proteins that pathogenic bacteria secrete upon infecting their plant host. Inside a plant nucleus, TALEs can act as transcriptional activators triggering processes favorable to the pathogen. Apart from their benefits to a pathogen, TALEs bring along interesting features that have also proven beneficial to synthetic biology applications.
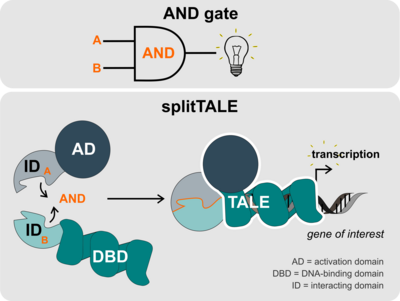
TALEs are equipped with a DNA-binding domain that consists of amino acid repeats each of which specifically binds a DNA base. Furthermore, TALEs have an activation domain that functions in activating gene expression. In the past, their DNA-binding and activation domain have been extensively engineered in order for TALEs to bind specific DNA sequences and there exert desired functions ranging from repression to DNA cleavage.
Inspired by other existing AND-gates in the synthetic biology toolset, Schreiber and coworkers have now developed one specifically for application in plants. For that, they split up a TALE’s activation and DNA-binding domain – thus calling their system “splitTALE”. Each domain was then fused to protein domains that are known to interact. Given those two interacting protein domains would come into close proximity, the TALE activation and DNA-binding domains would form a functional TALE capable to activate transcription of, e.g., a TALE-controlled reporter gene or another gene of interest. Two interacting components, therefore, have to be present for such a genetic AND-gate to produce the desired output.
To reliably trigger reporter expression in plants, the researchers thoroughly optimized the splitTALE system. First, they reduced background activity and improved signal-to-noise ratio of the reporter by introducing several truncations to the DNA-binding component and by finding the best positioning of a nuclear localization signal. Also, different activation domains were assessed for optimal reporter output. Next, they tested different interaction domains to exclude that cross-interaction occurs and to ensure specificity of the ‘AND’ gate. With the optimized splitTALE at hand, Schreiber et al. finally applied it to control production of the high-value terpene Z-abienol in otherwise Z-abienol-free plants, thereby demonstrating the potential of splitTALE as a useful asset in the metabolic engineering toolbox.
Reminiscent of protein-protein-interaction assays like bimolecular fluorescence complementation (also known as splitYFP), splitTALE offers a more reliable, quantifiable in planta tool for protein-protein-interaction analysis due to its virtually absent background activity and versatility with regard to reporter gene choice. Prospective uses as AND gate include targeting gene expression to overlapping expression domains for example. Another advantage of the splitTALE system is the adaptability of its DNA-binding domain to bind sequences of interest including promoter regions of endogenous genes. Since the splitTALE system has been integrated with the GoldenGate modular cloning system, scientists can easily apply and customize it for their applications of interest.
Original Publication: Tom Schreiber, Anja Prange, Tina Hoppe, Alain Tissier (2019). Split-TALE: A TALE-based two-component system for synthetic biology applications in planta, Plant Physiology, pp.01218.2018; DOI: 10.1104/pp.18.01218